Part:BBa_K3468088:Design
PETase A202C&E231C
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Design Notes
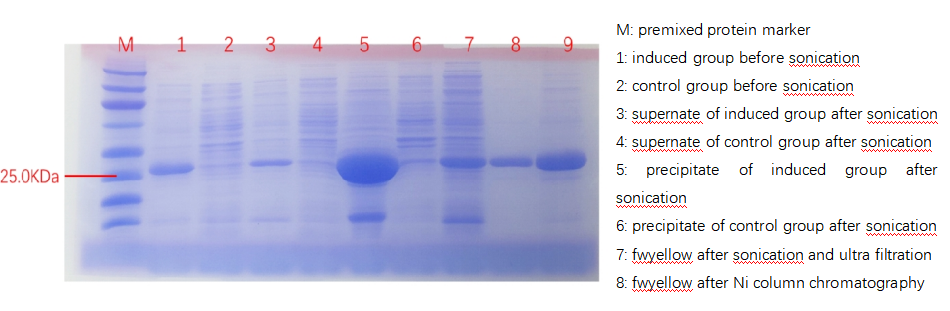
It should be noted that not all engineered disulfides have provided an increase in stability, as there are a number of reports of destabilizing disulfides. Given the mixed outcomes, disulfide engineering studies would benefit greatly from computational tools that not only identify novel disulfides that are likely to form, but also indicate whether a disulfide is likely to confer an increase in stability. To introduce disulfide bonds in PETase, we used computer software Disulfide by Design (DbD) and Modelling of Disulfide Bonds in Proteins (MODIP) to identify mutation sites where the disulfide bonds were most likely to form. MODIP The DbD2 algorithm can accurately and rapidly estimate the Cβ–Sγ–Sγ–Cβ (χ3) torsion angle which represents rotation of the Cβ atoms, and analyse B-factors of candidate disulfide regions to help identify those that may confer increased stability to the protein. In MODIP, to evaluate the modeled disulfide, a gradation scheme was used to rank the predicted disulfides based on certain stereochemical parameters (dihedral angles and S–S bond distance). Thus, a disulfide bridge modeled with dihedral angles within an ideal range of values, typical of naturally occurring disulfide bridges, was assigned grade ‘A’. A site which was geometrically suitable for formation of the S–S covalent bond, but would have a somewhat distorted stereochemistry, was assigned grade ‘B’, and sites which were simply close enough in space potentially to allow the formation of the disulfide bond were assigned grade ‘C’. Through these two programs, we screened for potential sites where mutations form disulfide bonds, A202C-E231C being one of the possible sites (as shown in Fig 1)
Source
Ideonalla sakaiensis
References
[1] Craig and Dombkowski: Disulfide by Design 2.0: a web-based tool for disulfide engineering in proteins. BMC Bioinformatics 2013 14:346. [2] Hein J.Wijma, Robert J.Floor, Peter A.Jekel1,David Baker, Siewert J.Marrink and Dick B.Janssen. Computationally designed libraries for rapid enzyme stabilization. Protein Engineering, Design & Selection vol. 27 no. 2 pp. 49– 58, 2014. [3] Vardhan S.Dani, C.Ramakrishnan and Raghavan Varadarajan. MODIP revisited: re-evaluation and refinement of an automated procedure for modeling of disulfide bonds in proteins. Protein Engineering vol.16 no.3 pp.187–193, 2003.